High-throughput pharmaceutical screening and structure based inhibitor development
High-throughput pharmaceutical screening and structure based inhibitor development
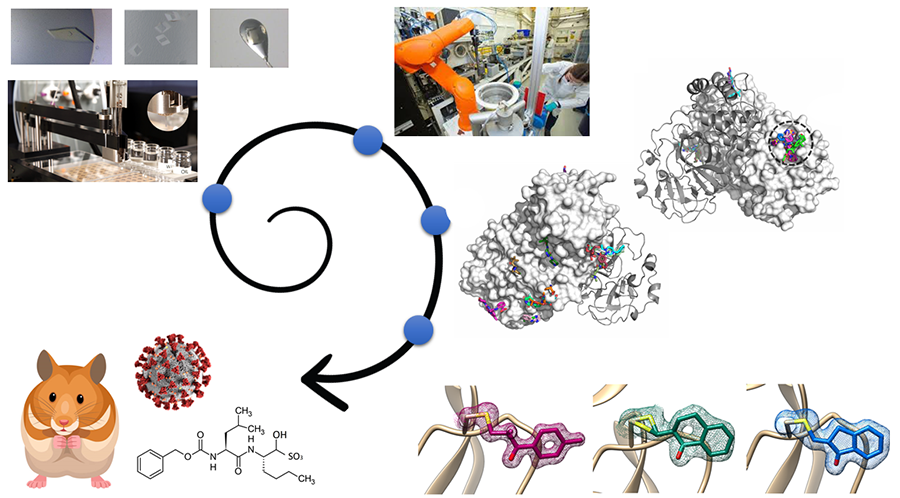
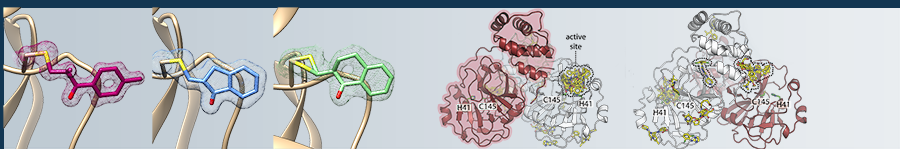
Structure-based drug development is among the most powerful techniques in modern drug discovery. Here, based on the three-dimensional structure of drug targets, for example proteins essential for viral replication or host cell entry, different binders are proposed typically by computational predictions or initial considerations of compound properties and then tested for the binding or even inhibition in biochemical and biophysical assays. Subsequently or in parallel to screening by assays, the pre-identified compounds or a larger library of compounds are investigated by means of X-ray crystallography, allowing to understand the mode of binding to the protein and relation of structure and function. Based on high-resolution structural data the compounds are then typically iteratively optimized to obtain higher binding affinities and inhibition of the protein function, improve the specificity or reduce toxicity. Our screening and compound optimization approaches are supported by complementary methods such as mass spectrometry. This process of structure-based drug development starting with the selection of a target protein and its 3D-structure determination and ideally ending with (pre-)clinical testing of promising compounds typically takes several years. In order to make this process more efficient we develop and continously improve high-throughput X-ray methods including the development of new hardware, such as multi crystal sample holders and robotics, and highly automated data collection routines. Furthermore, we collaborate with partners to improve data analysis by including AI methods. Altogether, the advanced infrastructure available on the DESY campus and in the Hamburg area enables us to rapidly initiate and eficientally perform such drug discovery projects in short time. At present, our investigations focus on finding new drug candidates against several highly relevant SARS-CoV-2 related proteins but also against proteins from other emerging RNA viruses, that are listed as potential candidates for a next pandemic.
Selected References
1. Günther, S.*, Reinke, P.Y.A.* et al. (2021), X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease. Science 372 (6542). DOI: 10.1126/science.abf7945
2. Ewert, W.*, Günther, S.* et al. (2022), Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease. Front Chem 10:832431.DOI: 10.3389/fchem.2022.832431. eCollection 2022
3. Srinivasan V et al. (2022), Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease. Commun Biol. 11;5(1):805. DOI: 10.1038/s42003-022-03737-7